Chapter 8 Data frames
(R data frames)
8.1 Overview
8.1.1 Abstract:
Introduction to data frames: how to create, and modify them and how to retrieve data.
8.1.2 Objectives:
This unit will:
- introduce R data frames;
- cover a number of basic operations.
8.1.3 Outcomes:
After working through this unit you:
- know how to create and manipulate data frames;
- can extract rows, columns, and append new data rows;
8.1.4 Deliverables:
Time management: Before you begin, estimate how long it will take you to complete this unit. Then, record in your course journal: the number of hours you estimated, the number of hours you worked on the unit, and the amount of time that passed between start and completion of this unit.
Journal: Document your progress in your Course Journal. Some tasks may ask you to include specific items in your journal. Don't overlook these.
Insights: If you find something particularly noteworthy about this unit, make a note in your insights! page.
8.1.5 Prerequisites
8.2 Data frames
8.3 Task 16 - Basic operations
Load the R-Exercise_BasicSetup project in RStudio if you don't already have it open. Type init() as instructed after the project has loaded. Continue below.
Data frames are probably the most important type of data object for bioinformatics in R; they emulate our mental model of data in a spreadsheet and can be used to implement entity-relationship datamodels.
Usually the result of reading external data from an input file is a data frame. The file below is included with the R-Exercise-BasicSetup project files - it is called plasmidData.tsv, and you can click on it in the Files Pane to open and inspect it.
This data set uses tabs as column separators and it has a header line. Similar files can be exported from Excel or other spreadsheet programs.
Read this as a data frame as follows:
( plasmidData <- read.table(
file.path("data_files","plasmidData.tsv"),
sep="\t",
header=TRUE,
stringsAsFactors = FALSE) )## Name Size Marker Ori Sites
## 1 pUC19 2686 Amp ColE1 EcoRI, SacI, SmaI, BamHI, XbaI, PstI, HindIII
## 2 pBR322 4361 Amp, Tet ColE1 EcoRI, ClaI, HindIII
## 3 pACYC184 4245 Tet, Cam p15A ClaI, HindIIIobjectInfo(plasmidData)## object contents: Name Size Marker Ori Sites
## 1 pUC19 2686 Amp ColE1 EcoRI, SacI, SmaI, BamHI, XbaI, PstI, HindIII
## 2 pBR322 4361 Amp, Tet ColE1 EcoRI, ClaI, HindIII
## 3 pACYC184 4245 Tet, Cam p15A ClaI, HindIII
##
## structure of object:
## 'data.frame': 3 obs. of 5 variables:
## $ Name : chr "pUC19" "pBR322" "pACYC184"
## $ Size : int 2686 4361 4245
## $ Marker: chr "Amp" "Amp, Tet" "Tet, Cam"
## $ Ori : chr "ColE1" "ColE1" "p15A"
## $ Sites : chr "EcoRI, SacI, SmaI, BamHI, XbaI, PstI, HindIII" "EcoRI, ClaI, HindIII" "ClaI, HindIII"
##
## attributes:## $names
## [1] "Name" "Size" "Marker" "Ori" "Sites"
##
## $class
## [1] "data.frame"
##
## $row.names
## [1] 1 2 3Note the argument stringsAsFactors = FALSE. If this is TRUE instead, R will convert all strings in the input to factors and this may lead to problems. Make it a habit to turn this behaviour off, you can always turn a column of strings into factors when you actually mean to have factors.
You can view the data frame contents by clicking on the spreadsheet icon behind its name in the Environment Pane.
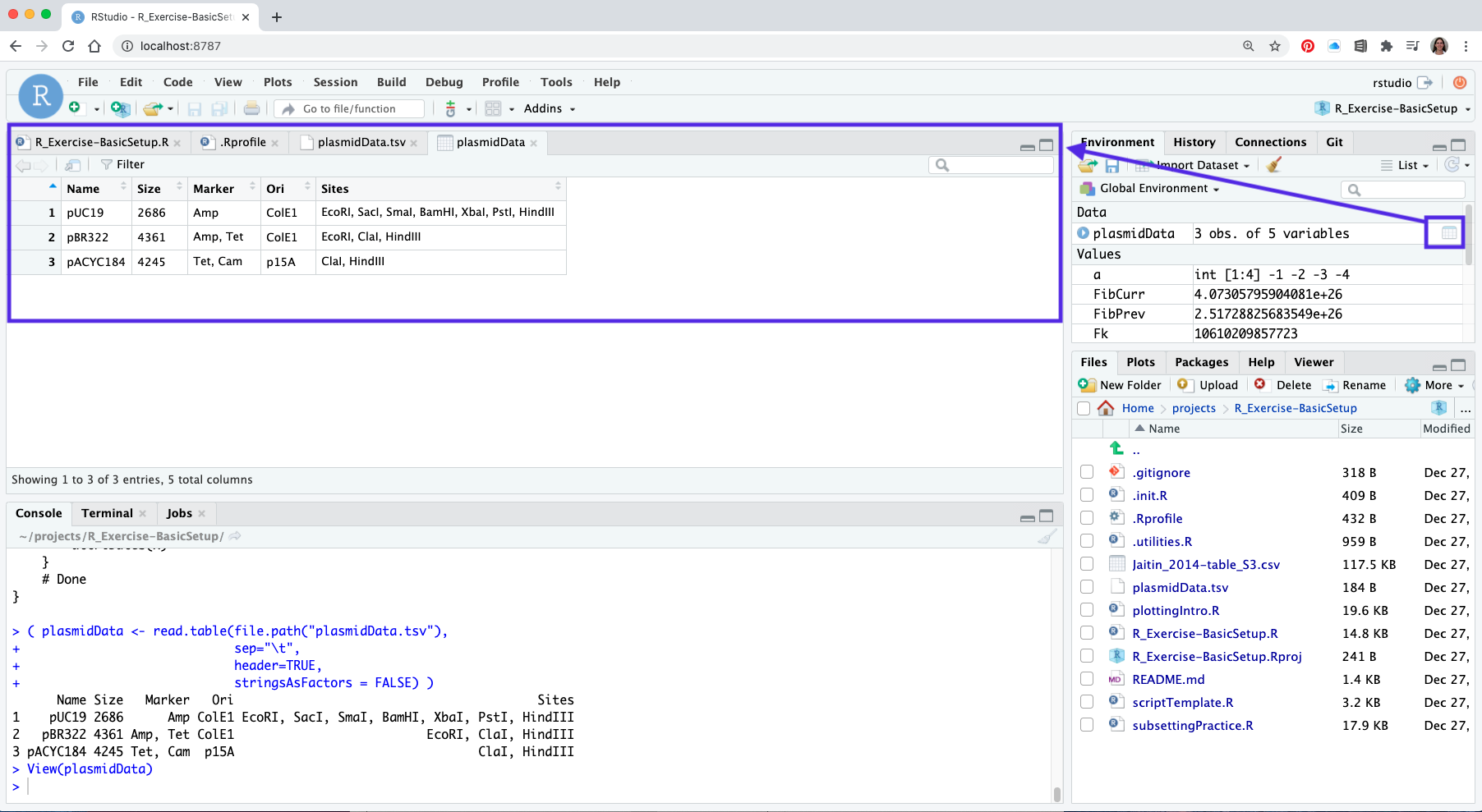
8.4 Basic operations
Here are some basic operations with the data frame. Try them and experiment. If you break it by mistake, you can just recreate it by reading the source file again:
use column 1 as rownames
rownames(plasmidData) <- plasmidData[ , 1] nrow(plasmidData)## [1] 3ncol(plasmidData)## [1] 5objectInfo(plasmidData)## object contents: Name Size Marker Ori
## pUC19 pUC19 2686 Amp ColE1
## pBR322 pBR322 4361 Amp, Tet ColE1
## pACYC184 pACYC184 4245 Tet, Cam p15A
## Sites
## pUC19 EcoRI, SacI, SmaI, BamHI, XbaI, PstI, HindIII
## pBR322 EcoRI, ClaI, HindIII
## pACYC184 ClaI, HindIII
##
## structure of object:
## 'data.frame': 3 obs. of 5 variables:
## $ Name : chr "pUC19" "pBR322" "pACYC184"
## $ Size : int 2686 4361 4245
## $ Marker: chr "Amp" "Amp, Tet" "Tet, Cam"
## $ Ori : chr "ColE1" "ColE1" "p15A"
## $ Sites : chr "EcoRI, SacI, SmaI, BamHI, XbaI, PstI, HindIII" "EcoRI, ClaI, HindIII" "ClaI, HindIII"
##
## attributes:## $names
## [1] "Name" "Size" "Marker" "Ori" "Sites"
##
## $class
## [1] "data.frame"
##
## $row.names
## [1] "pUC19" "pBR322" "pACYC184"assign one row to a variable. This is also a data frame! One row. It has to be, because it contains elements of type chr and of type int!
x <- plasmidData[2, ]
objectInfo(x) ## object contents: Name Size Marker Ori Sites
## pBR322 pBR322 4361 Amp, Tet ColE1 EcoRI, ClaI, HindIII
##
## structure of object:
## 'data.frame': 1 obs. of 5 variables:
## $ Name : chr "pBR322"
## $ Size : int 4361
## $ Marker: chr "Amp, Tet"
## $ Ori : chr "ColE1"
## $ Sites : chr "EcoRI, ClaI, HindIII"
##
## attributes:## $names
## [1] "Name" "Size" "Marker" "Ori" "Sites"
##
## $row.names
## [1] "pBR322"
##
## $class
## [1] "data.frame"retrieve one row: different syntax, same thing
plasmidData["pBR322", ] ## Name Size Marker Ori Sites
## pBR322 pBR322 4361 Amp, Tet ColE1 EcoRI, ClaI, HindIIIretrieve one column - two different methods
plasmidData[ , 2] ## [1] 2686 4361 4245plasmidData[ , "Size"] ## [1] 2686 4361 4245remove one row
plasmidData <- plasmidData[-2, ]
objectInfo(plasmidData)## object contents: Name Size Marker Ori
## pUC19 pUC19 2686 Amp ColE1
## pACYC184 pACYC184 4245 Tet, Cam p15A
## Sites
## pUC19 EcoRI, SacI, SmaI, BamHI, XbaI, PstI, HindIII
## pACYC184 ClaI, HindIII
##
## structure of object:
## 'data.frame': 2 obs. of 5 variables:
## $ Name : chr "pUC19" "pACYC184"
## $ Size : int 2686 4245
## $ Marker: chr "Amp" "Tet, Cam"
## $ Ori : chr "ColE1" "p15A"
## $ Sites : chr "EcoRI, SacI, SmaI, BamHI, XbaI, PstI, HindIII" "ClaI, HindIII"
##
## attributes:## $names
## [1] "Name" "Size" "Marker" "Ori" "Sites"
##
## $row.names
## [1] "pUC19" "pACYC184"
##
## $class
## [1] "data.frame"add it back at the end
plasmidData <- rbind(plasmidData, x)
objectInfo(plasmidData)## object contents: Name Size Marker Ori
## pUC19 pUC19 2686 Amp ColE1
## pACYC184 pACYC184 4245 Tet, Cam p15A
## pBR322 pBR322 4361 Amp, Tet ColE1
## Sites
## pUC19 EcoRI, SacI, SmaI, BamHI, XbaI, PstI, HindIII
## pACYC184 ClaI, HindIII
## pBR322 EcoRI, ClaI, HindIII
##
## structure of object:
## 'data.frame': 3 obs. of 5 variables:
## $ Name : chr "pUC19" "pACYC184" "pBR322"
## $ Size : int 2686 4245 4361
## $ Marker: chr "Amp" "Tet, Cam" "Amp, Tet"
## $ Ori : chr "ColE1" "p15A" "ColE1"
## $ Sites : chr "EcoRI, SacI, SmaI, BamHI, XbaI, PstI, HindIII" "ClaI, HindIII" "EcoRI, ClaI, HindIII"
##
## attributes:## $names
## [1] "Name" "Size" "Marker" "Ori" "Sites"
##
## $row.names
## [1] "pUC19" "pACYC184" "pBR322"
##
## $class
## [1] "data.frame"add a new row from scratch:
plasmidData <- rbind(plasmidData,
data.frame(Name = "pMAL-p5x", Size = 5752,
Marker = "Amp",Ori = "pMB1",
Sites = "SacI, AvaI, ..., HindIII",
stringsAsFactors = FALSE))
objectInfo(plasmidData)## object contents: Name Size Marker Ori
## pUC19 pUC19 2686 Amp ColE1
## pACYC184 pACYC184 4245 Tet, Cam p15A
## pBR322 pBR322 4361 Amp, Tet ColE1
## 1 pMAL-p5x 5752 Amp pMB1
## Sites
## pUC19 EcoRI, SacI, SmaI, BamHI, XbaI, PstI, HindIII
## pACYC184 ClaI, HindIII
## pBR322 EcoRI, ClaI, HindIII
## 1 SacI, AvaI, ..., HindIII
##
## structure of object:
## 'data.frame': 4 obs. of 5 variables:
## $ Name : chr "pUC19" "pACYC184" "pBR322" "pMAL-p5x"
## $ Size : num 2686 4245 4361 5752
## $ Marker: chr "Amp" "Tet, Cam" "Amp, Tet" "Amp"
## $ Ori : chr "ColE1" "p15A" "ColE1" "pMB1"
## $ Sites : chr "EcoRI, SacI, SmaI, BamHI, XbaI, PstI, HindIII" "ClaI, HindIII" "EcoRI, ClaI, HindIII" "SacI, AvaI, ..., HindIII"
##
## attributes:## $names
## [1] "Name" "Size" "Marker" "Ori" "Sites"
##
## $row.names
## [1] "pUC19" "pACYC184" "pBR322" "1"
##
## $class
## [1] "data.frame"8.5 Task 17 - modify data frame
The rowname of the new row of plasmidData is now "1". It should be "pMAL-p5x". Fix this.
8.6 Self-evaluation
8.7 Further reading, links and resources
If in doubt, ask!
If anything about this learning unit is not clear to you, do not proceed blindly but ask for clarification. Post your question on the course mailing list: others are likely to have similar problems. Or send an email to your instructor.
Author: Boris Steipe boris.steipe@utoronto.ca
Created: 2017-08-05
Modified: 2018-05-05
Version: 1.0.1
Version history:
1.0.1 Maintenance
1.0 Completed to first live version
0.1 Material collected from previous tutorial
8.7.1 Updated Revision history
| Revision | Author | Date | Message |
|---|---|---|---|
| 5152496 | Ruth Isserlin | 2019-12-25 | Fixed issue with task numbering because of a remove all elements call in the 2nd chapter |
| fab47ae | Ruth Isserlin | 2019-12-24 | initial check in of R basics book |